optiBET is a shell script to perform brain-extractions even in patients brain, where extensive pathology is present. The script has now been tested on a set of more than 70 patient brains, and it performs better than all available tools (see figure below).
Lutkenhoff ES, Rosenberg M, Chiang J, Zhang K, Pickard JD, Owen AM, Monti MM. (2014) Optimized Brain Extraction for Pathological Brains (optiBET). PLoS ONE 9(12): e115551. doi:10.1371/journal.pone.0115551
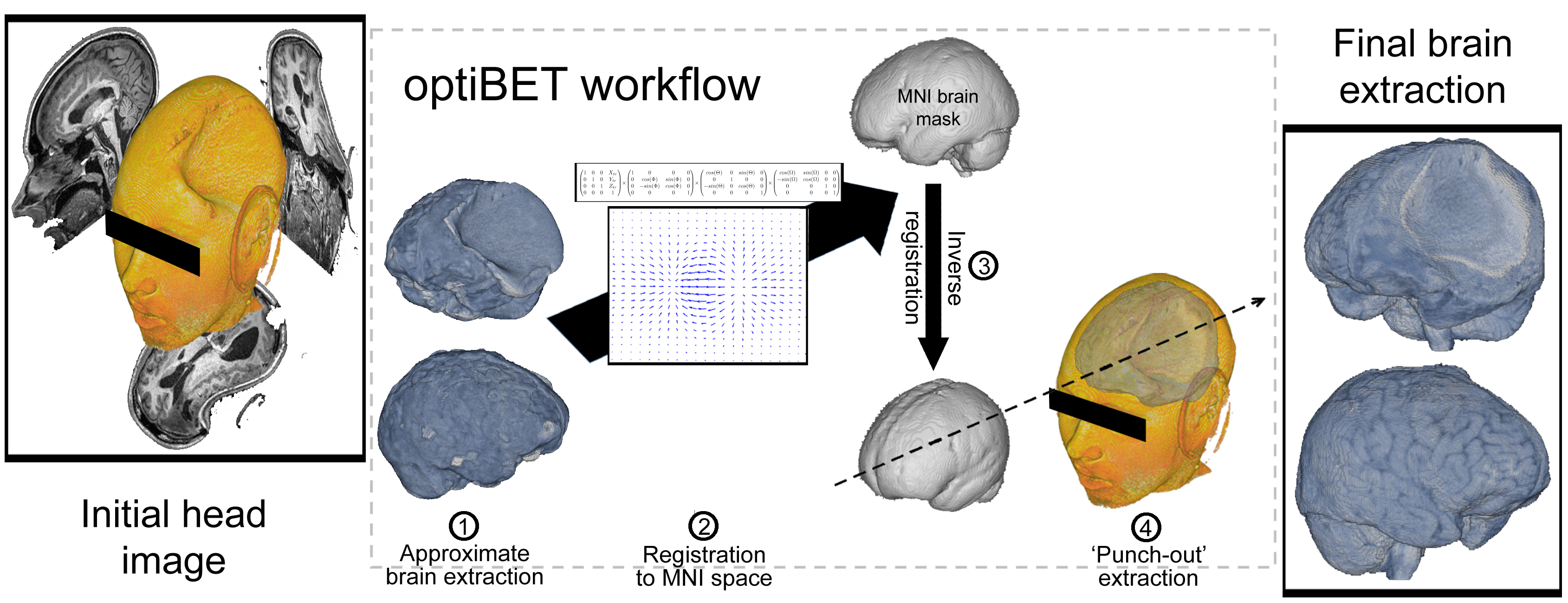
INSTALLATION: just put the script in your path (and if needed make it executable; e.g., “chmod 755 optibet.sh”). The only requirement for the default options is to have FSL installed. If you want to use the AFNI option (which in our evaluation is sub-optimal!) then you will need both FSL and AFNI installed.
USAGE: sh optiBET.sh -i <input_image> -options
Type sh optiBET.sh for a full list of options
-f script uses FSL for initial extraction (default)
-a script uses AFNI for initial extraction
-o script uses MNI152_T1_1mm_brain_mask.nii.gz for mask (default)
-t script uses MNI152_T1_2mm_brain_mask.nii.gz for mask
-g script uses avg152T1_brain.nii.gz for mask
-d use debug mode (will NOT delete intermediate files)
*script requires proper installation of FSL and AFNIinput image should be in standard orientation; use .nii.gz image for input; outputs binarized brain-extraction mask, saved as: <input_image>_optiBET_brain_mask.nii.gz, and full-intensity brain-extraction, saved as: <input_image>_optiBET_brain.nii.gz
* Depending on your shell and configuration (e.g. some Ubuntu installations) , please start optiBET by typing:
sh optiBET.sh -i input OR
bash optiBET.sh -i input
Comparison data
(“Manual” is the manually extracted ‘golden-standard’ to which all other extractions are compared. RBX: Robex; 3dSS: 3dSkullStrip; us: useskull;)
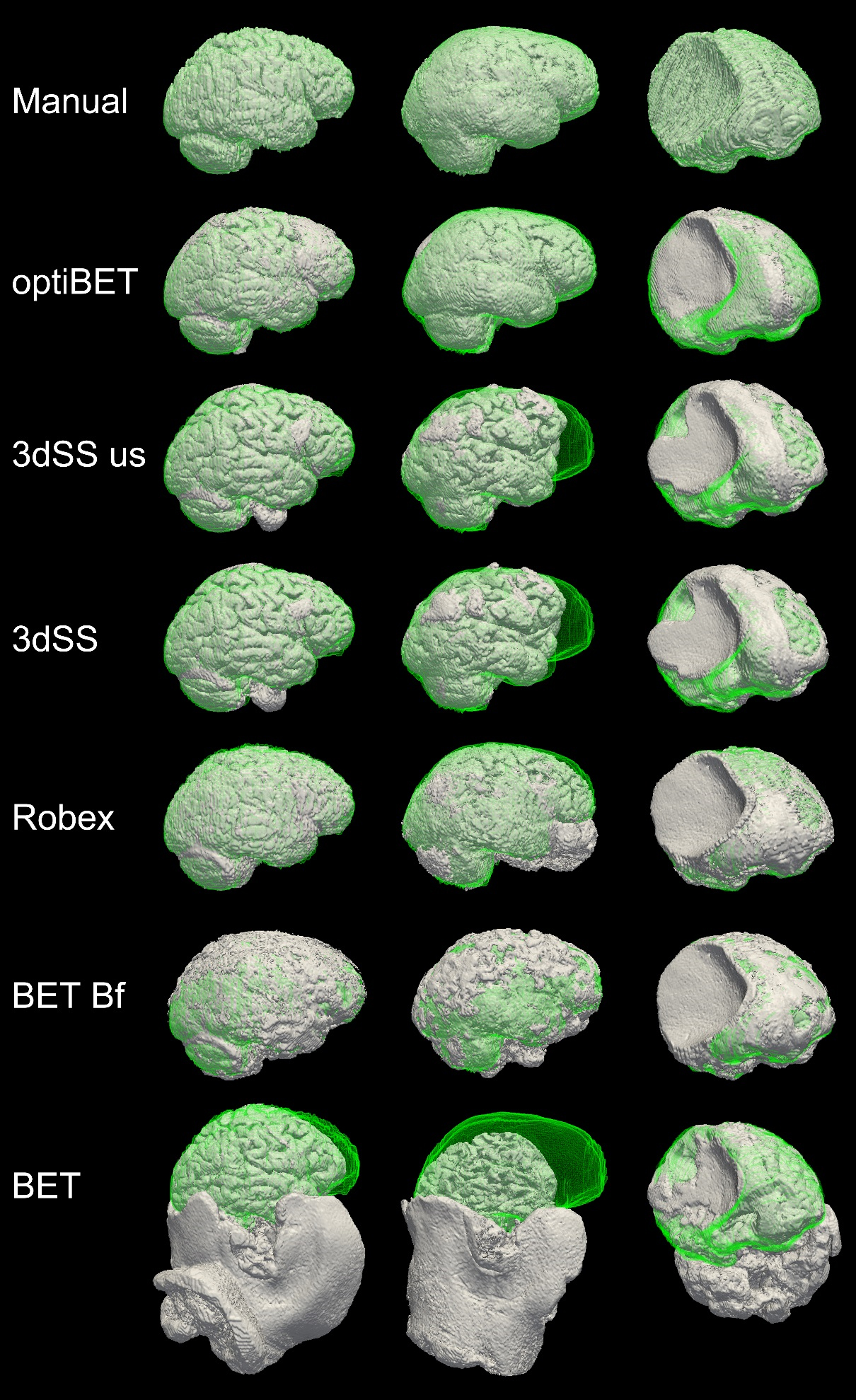
Sample extraction results. Rendering of brain extractions obtained from different algorithms/options (shown in grey) compared to the manually-traced benchmark (shown in green wireframe) for three sample patients with different degrees of brain pathology (little, medium, and high, for the left, middle and right columns respectively). The first row depicts the rendering of the benchmark volume (in grey) and the benchmark wireframe to illustrate a case of “perfect fit.” (Renderings obtained with ParaView; Hendrson, 2004). Areas where no green is visible indicate regions in which the brain extraction was overly permissive, thus failing to remove some non-brain tissue (e.g., row 5, column 2, in the figure). Areas where only the green wireframe is visible (e.g., rows 3 and 4, column 2), and no grey area is, indicate regions in which the brain extraction was overly conservative, removing brain tissue.
